Execute thousands of computational tasks to analyse large datasets
HADDOCK helps scientists to advance COVID-19 research
Talking about impact in the context of the novel COVID-19 research is still difficult in terms of finding a vaccine or any other solution to eliminate the virus. That is not to say that there isn’t any progress being made, by plenty of researchers, many of them benefiting from open science initiatives.
One example of these initiatives is HADDOCK – High Ambiguity Driven protein-protein DOCKing – developed at Utrecht University and operated as a core software in the BioExcel Center of Excellence to support COVID-19 researchers.
The HADDOCK platform supported the drug repurposing effort by screening over 2000 chemical compounds against 3 COVID-19 related targets in up to 8 days using EGI High-Throughput Compute resources.
HADDOCK supports complex simulations to generate 3D models revealing how virus proteins interact with human ones, or to dock small molecules to targets such as the SARS-CoV-2 protease.
This potential drug target plays an essential role in processing the polyproteins that are translated from the viral RNA during replication. Targeting it and other viral proteins such as the RNA polymerase with small molecules could make for effective anti-coronaviral drug cocktails (as is done for example for HIV).
Due to the urgency, researchers made use of a strategy that repurposes existing and already approved drugs toward new diseases. The HADDOCK WeNMR platform supported this drug repurposing effort by screening over 2000 chemical compounds against three COVID-19 related targets in up to 8 days using EGI High-Throughput Compute resources.
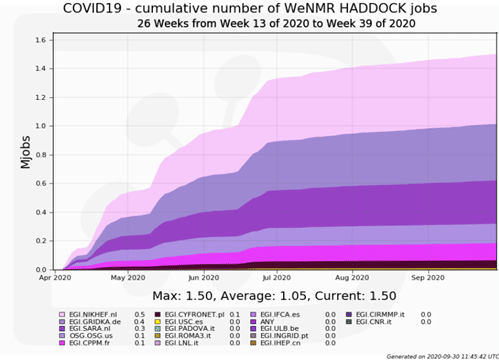
Thanks to the synergies and cooperation already established between EGI, the Open Science Grid (USA) and various high energy physics sites that committed to support COVID-19 related research, HADDOCK was able to more than double its processing capacity, serving on average 550 active users per months, who submitted over 11,000 simulations related to COVID-19 (the equivalent of ~1.5 million HTC jobs, ~2.7 million CPU hours) on the EGI and OSC grid resources over the months of April to September 2020.
The initial results have revealed interesting compounds, some of which are already under clinical trials, supporting the validity of the screening methodology. A selection of these compounds is currently being experimentally tested by the Veterinary Faculty at Utrecht University.
In order to monitor COVID-19 related runs, the portal allows tagging of submission as COVID-19. Since this monitoring started, over 11.000 runs – not counting the drug repurposing screen – were processed to date. This tagging also allows to target the HTC jobs to sites specifically supporting this research. Another great result is to see the international mobilisation of additional resources to support HADDOCK jobs from both European, often high energy physics sites like Nikhef (Netherlands), CPPM (France), KIT (Germany) and USC-LCG2 (Spain), and the Open Science Grid (USA).
